By definition, dictionary are not sorted (to speed up access). Let us consider the following dictionary, which stores the age of several persons as values:
d = {"Pierre": 42, "Anne": 33, "Zoe": 24} |
d = {"Pierre": 42, "Anne": 33, "Zoe": 24}
If you want to sort this dictionary by values (i.e., the age), you must use another data structure such as a list, or an ordered dictionary.
Use the sorted function and operator module
import operator
sorted_d = sorted(d.items(), key=operator.itemgetter(1)) |
import operator
sorted_d = sorted(d.items(), key=operator.itemgetter(1))
Sorted_d is a list of tuples sorted by the second element in each tuple. Each tuple contains the key and value for each item found in the dictionary. If you look at the content of this variable, you should see:
[ ('Zoe', 24), ('Anne', 33), ('Pierre', 42)] |
[ ('Zoe', 24), ('Anne', 33), ('Pierre', 42)]
Use the sorted function and lambda function
If you do not want to use the operator module, you can use a lambda function:
sorted_d = sorted(d.items(), key=lambda x: x[1])
# equivalent version
# sorted_d = sorted(d.items(), key=lambda (k,v): v) |
sorted_d = sorted(d.items(), key=lambda x: x[1])
# equivalent version
# sorted_d = sorted(d.items(), key=lambda (k,v): v)
The computation time is of the same order of magnitude as with the operator module. Would be interesting to test on large dictionaries.
Use the sorted function and return an ordered dictionary
In the previous methods, the returned objects are list of tuples. So we do not have a dictionary anymore. You can use an OrderedDict if you prefer:
>>> from collections import OrderedDict
>>> dd = OrderedDict(sorted(d.items(), key=lambda x: x[1]))
>>> print(dd)
OrderedDict([('Pierre', 24), ('Anne', 33), ('Zoe', 42)]) |
>>> from collections import OrderedDict
>>> dd = OrderedDict(sorted(d.items(), key=lambda x: x[1]))
>>> print(dd)
OrderedDict([('Pierre', 24), ('Anne', 33), ('Zoe', 42)])
Use sorted function and list comprehension
Another method consists in using list comprehension and use the sorted function on the tuples made of (value, key).
sorted_d = sorted((value, key) for (key,value) in d.items()) |
sorted_d = sorted((value, key) for (key,value) in d.items())
Here the output is a list of tuples where each tuple contains the value and then the key:
[(24, 'Pierre'), (33, 'Anne'), (42, 'Zoe')] |
[(24, 'Pierre'), (33, 'Anne'), (42, 'Zoe')]
A note about Python 3.6 native sorting
In previous version on this post, I wrote that “In Python 3.6, the iteration through a dictionary is sorted.”. This is wrong. What I meant is that in Python 3.6 dictionary keeps insertion sorted.
It means that if you insert your items already sorted, the new python 3.6 implementation will be this information. Therefore, there is no need to sort the items anymore. Of course, if you insert the items randomly, you will still need to use one of the method mentioned above.
For instance, taking care of the age, we now create our list as follows (sorting by ascending age):
d = {("Zoe": 24)}
d.update({'Anne': 33})
d.update({'Pierre': 42}) |
d = {("Zoe": 24)}
d.update({'Anne': 33})
d.update({'Pierre': 42})
Now you can iterate through the items and they will be in the same order as in the creation of the dictionary. So you can just create a list from your items very easily:
list(d.items())
Out[15]: [('Zoe', 24), ('Anne', 33), ('Pierre', 42)] |
list(d.items())
Out[15]: [('Zoe', 24), ('Anne', 33), ('Pierre', 42)]
Benchmark
Here is a quick benchmark made using the small dictionary from the above examples. Would be interesting to redo the test with a large dictionary.
What you can see is that the native Python dictionary sorting is pretty cool followed by the combination of the lambda + list comprehension method. Overall using one of these methods would be equivalent though (factor 2/3 at most).
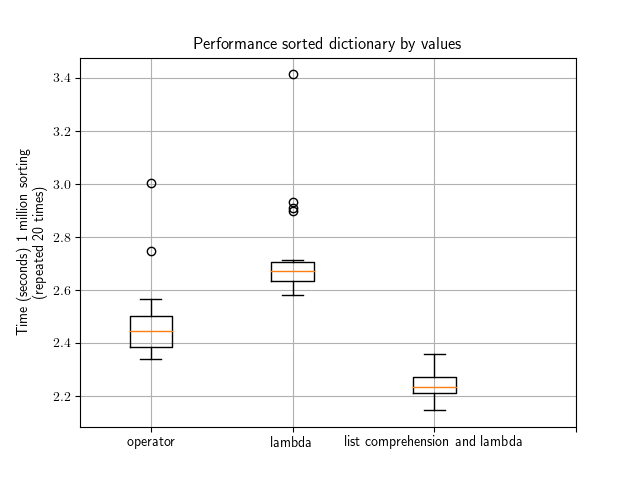
This image was created with the following code.
import operator
import pylab
from easydev import Timer
times1, times2, times3 = [], [], []
pylab.clf()
d = {"Pierre": 42, "Anne": 33, "Zoe": 24}
for j in range(20):
N = 1000000
with Timer(times3):
for i in range(N):
sorted_d = sorted((key, value) for (key,value) in d.items())
with Timer(times2):
for i in range(N):
sorted_d = sorted(d.items(), key=lambda x: x[1])
with Timer(times1):
for i in range(N):
sorted_d = sorted(d.items(), key=operator.itemgetter(1))
print(j)
pylab.boxplot([times1, times2, times3])
pylab.xticks([1,2,3], ["operator", "lambda", "list comprehension and lambda"])
pylab.ylabel("Time (seconds) 1 million sorting \n (repeated 20 times)")
pylab.grid()
pylab.title("Performance sorted dictionary by values") |
import operator
import pylab
from easydev import Timer
times1, times2, times3 = [], [], []
pylab.clf()
d = {"Pierre": 42, "Anne": 33, "Zoe": 24}
for j in range(20):
N = 1000000
with Timer(times3):
for i in range(N):
sorted_d = sorted((key, value) for (key,value) in d.items())
with Timer(times2):
for i in range(N):
sorted_d = sorted(d.items(), key=lambda x: x[1])
with Timer(times1):
for i in range(N):
sorted_d = sorted(d.items(), key=operator.itemgetter(1))
print(j)
pylab.boxplot([times1, times2, times3])
pylab.xticks([1,2,3], ["operator", "lambda", "list comprehension and lambda"])
pylab.ylabel("Time (seconds) 1 million sorting \n (repeated 20 times)")
pylab.grid()
pylab.title("Performance sorted dictionary by values")